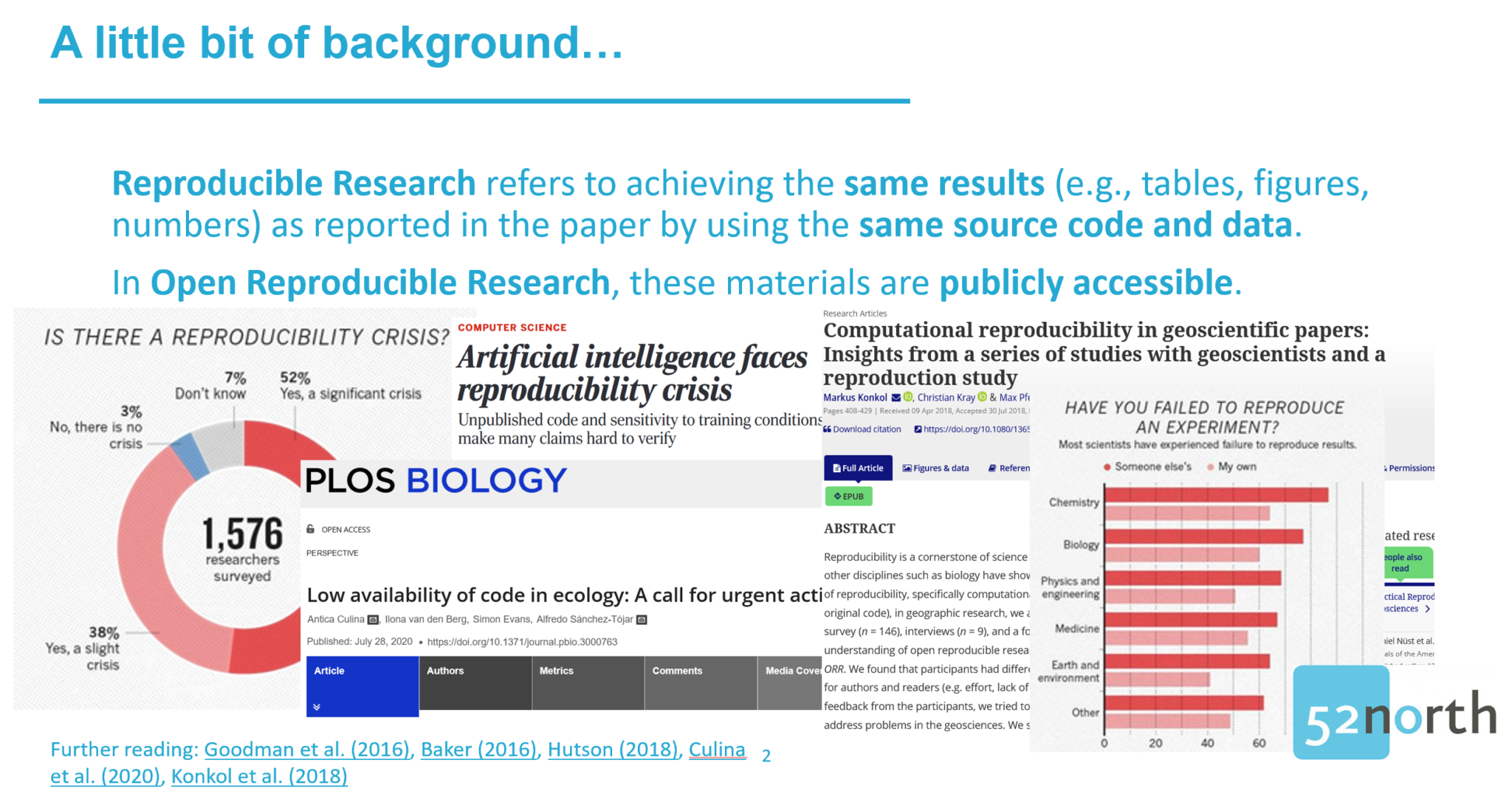
It’s Tuesday. 9pm. Silence in room 2. Typing. Whispering. Incredulous laughter. I am standing in front of 15 people staring at their screens, relieved to see that some of them struggle. Relieved? Exactly! But let’s take a step back.
We are at OpenGeoHub’s Summer School, an annual week-long event where experts on open source software and open data provide lectures for earth sciences researchers interested in improving their analysis and modelling frameworks. There was a strong focus on the programming languages R and Python, but Julia also had its slot and the geospatial content management system GeoNode was presented too. No need to say that 52°North is inherently interested in such an event and happy to help organize, support and contribute as part of the KI:STE project. Two of the many sessions were dedicated to open reproducible research (ORR). Reproducible Research refers to achieving the same results (e.g., tables, figures, numbers) as reported in the paper by using the same source code and data. In ORR, these materials are publicly accessible. ORR is an important requirement to achieve computational results that are fully transparent, verifiable, and reusable. It is otherwise hard for others to understand how the authors came to their conclusions, check whether the analysis was implemented correctly, and build on top of already existing work. Nevertheless, several studies revealed that ORR is not common [1, 2, 3, 4]. The materials are often unavailable or incomplete and even accessible source code and research data does not guarantee executable workflows. Several technical issues (e.g., wrong file directories or a different computational environment) can make it difficult for others to run the workflow on the own machine. To make it even more complicated: Even executable source code does not necessarily output the same figures as reported in the paper, e.g., because of a deviating software version.
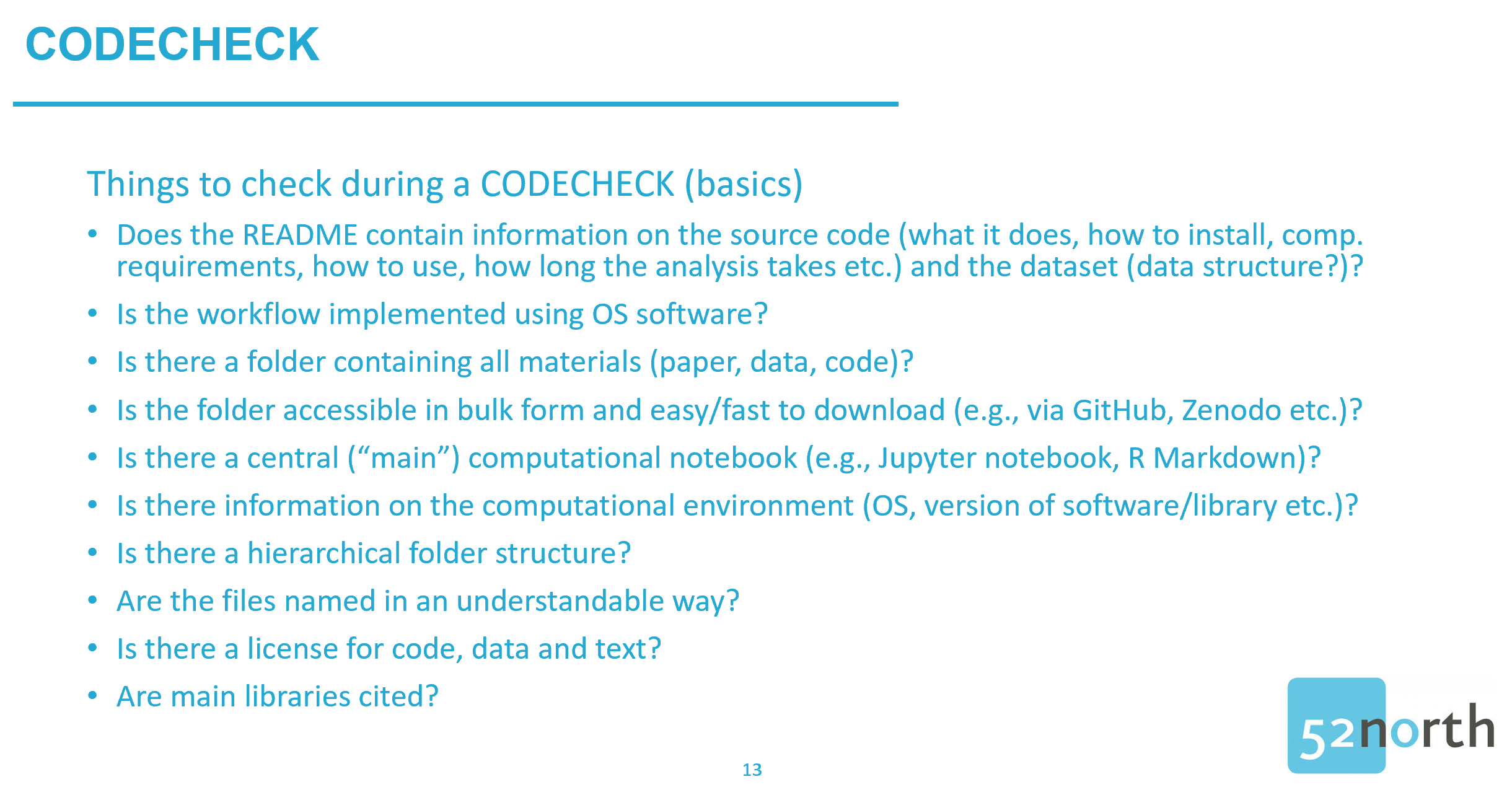
Carrying out studies to reveal reproducibility issues is helpful to raise awareness for unreproducible research results. However, reproducibility checks shouldn’t happen after the paper gets published. Wouldn’t it be better if such checks happened right before publication to make sure that there is no unpleasant surprise? It’s better, yes, but the peer review is already over and it would not be very nice to tell reviewers, who invested a substantial amount of work in checking the paper: „Well, thanks for reviewing, but we just found out that the results are not reproducible and we cannot publish the paper that you just spent several hours reviewing“. Sure, reviewers could do such a reproducibility check too, but aren’t they already overloaded with review requests? What if they don’t have the necessary technical skills? Ideally, reproducibility checks should be done at some point after the analysis is done and before submitting the paper for peer review. This approach would allow researchers to correct their workflow and save reviewers’ time. And that’s where CODECHECK comes into play. CODECHECK is a rather young initiative and was developed by Stephen Eglen and Daniel Nüst [5].

In a nutshell: The philosophy underlying CODECHECK is simple and can be summarized in a single question: “Was the code reproducible once for someone else?” Chances are good that if the code runs on one other computer, it will also run on several others. In a CODECHECK process, we have two roles: The author of the computational analysis and the Codechecker, who tries to run the analysis on their own machine. The process is composed of four steps. First, the author provides the Codechecker with access to the source code and data underlying the computational results in the paper. Second, the Codechecker tries to execute the analysis following the author’s written instructions (a README is a must-have!). Then, the Codechecker checks if the reproduced outputs resemble the figures, tables, and numbers reported in the manuscript. If this is not the case, the Codechecker and the author can check together if the differences are crucial and why they occur. If the code is not executable, the Codechecker can notify the author, ask for help, and then try it again. This process can have several iterations and may result in recommendations on how to improve the code before the author submits the manuscript for peer review. Third, in the case of a successful reproduction, the Codechecker can create a certificate including information on how the code was executed, what was checked, a copy of the reproduced outputs (i.e., figures, tables, and numbers), differences between original and reproduced outputs (and why they occur), and recommendations to improve the code. During the final step four, the Codechecker publishes the certificate on a repository, for example, Zenodo [6]. This will allow the author to cite the certificate in the supplementary materials section of the manuscript, thus giving credit to the Codechecker. The certificate shows reviewers that a CODECHECK was requested and carried out successfully.
Back to the Summer School: The morning session on reproducibility was dedicated to the theoretical concepts behind ORR, which technical issues make ORR difficult, and some practices to avoid these issues (see slides). In the evening, Benedikt Gräler and I planned a reproducibility hackathon to apply some of the lessons learned. Our idea was to let participants do a CODECHECK as described above (leaving out the steps to create a certificate). Honestly speaking, I had a few concerns. Many of the Summer School participants were experienced R and Python developers, some of them even authored software packages. So, what can I teach them? And who is actually willing to share materials? I was a bit worried that such a session might be a waste of time and leave the participants disappointed.
But which participants? It’s 8:10pm, the session started 10 minutes ago, and the room is almost empty. Anyway, it was worth a try…. But wait, several people come in, let’s wait a bit. A few minutes later, I am standing in front of 15 people providing them with a brief introduction to CODECHECK. Fortunately, five of them shared their GitHub repositories including source code and research data as use cases with the other participants.
What has motivated them to share their materials during the session? Diana Galindo stated:
“I consider that reproducibility is synonymous with good quality of a study and also allows progress towards new analyses on bases that are already running and it clearly saves time. […] it is a way to guarantee transparency in institutional processes and finally for the citizens.”
Another participant “[…] (correctly) assumed that other people looking at my materials could highlight some issues that I had not thought about before, and I would be able to learn something in the process.”
The other participants accessed the shared materials and, indeed, several of them struggled to reproduce the workflows. Reasons for that were manifold: For example, the code imported files using absolute instead of relative file paths; datasets were missing; source code files were not available or incomplete and so on. Was it mean to be relieved about these issues? Maybe, but I was just happy to see that the participants took something home, as one participant explained:
“[…] It also allowed me to look at my materials and how they are organized through the eyes of other people. As a result, I plan to apply a Codecheck to some of my work in the future.”
Martin Fleischmann (geopandas developer) learned “[…] that immediate reproducibility is one thing but the long-term reproducibility is another. We need to put more effort into ensuring that all materials can be easily used in years from now, not only when the paper gets published.”
I cannot speak for everyone, but I left the session with a happy face and some lessons learned:
- Participants of reproducibility hackathons can be late 😉.
- A reproducibility hackathon is worth the effort. There is always an issue in someone’s code.
- Letting others try to run the code can be a helpful lesson for the author.
Thanks to everyone who attended the session and special thanks to the volunteers who shared their materials with the other participants!
Leave a Reply